You can download the poster (PDF) at the downloadbox.
Tekstversion of the poster
Authors
Karin Elberse1, Laura Selva2, Gerlinde Pluister1, Natalia Peris2, Arie van der Ende3, Carmen Muñoz-Almagro2
- The Centre for Infectious Disease Control Netherlands (CIb), National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands
- Molecular Microbiology Department, University Hospital Sant Joan de Deu, Barcelona, Spain
- Reference Laboratory for Bacterial Meningitis, Academic Medical Center, Amsterdam, the Netherlands
Intoduction
In the era of global pneumococcal vaccination, selective
pressure on the pneumococcal population may lead to changes in its
composition. To monitor these alterations in the population,
genotyping tools for large-scale applications are essential. For
this, we compared the multiple-locus variable number tandem repeat
analysis (MLVA) with multilocus sequence typing (MLST) using 2
different pneumococcal populations, isolated from patients with
invasive disease from Catalonia and the Netherlands.
Method
The MLST and MLVA were performed as previously described
(Enright & Spratt, Microbiology 1998 and Elberse et al. PLoS
One 2011, http://www.mlva.net/). Pneumococcal
populations of 163 and
166 strains obtained in Catalan region in Spain and in the
Netherlands, respectively, were genotyped. The population included
consecutive pediatric strains from Catalan pediatric patients
(n=78) and all Dutch pediatric strains (n=71) of 2009-2012 and
consecutive strains in the age group >5 year from Catalonia
(n=85) and the Netherlands (n=95) of 2009-2012.
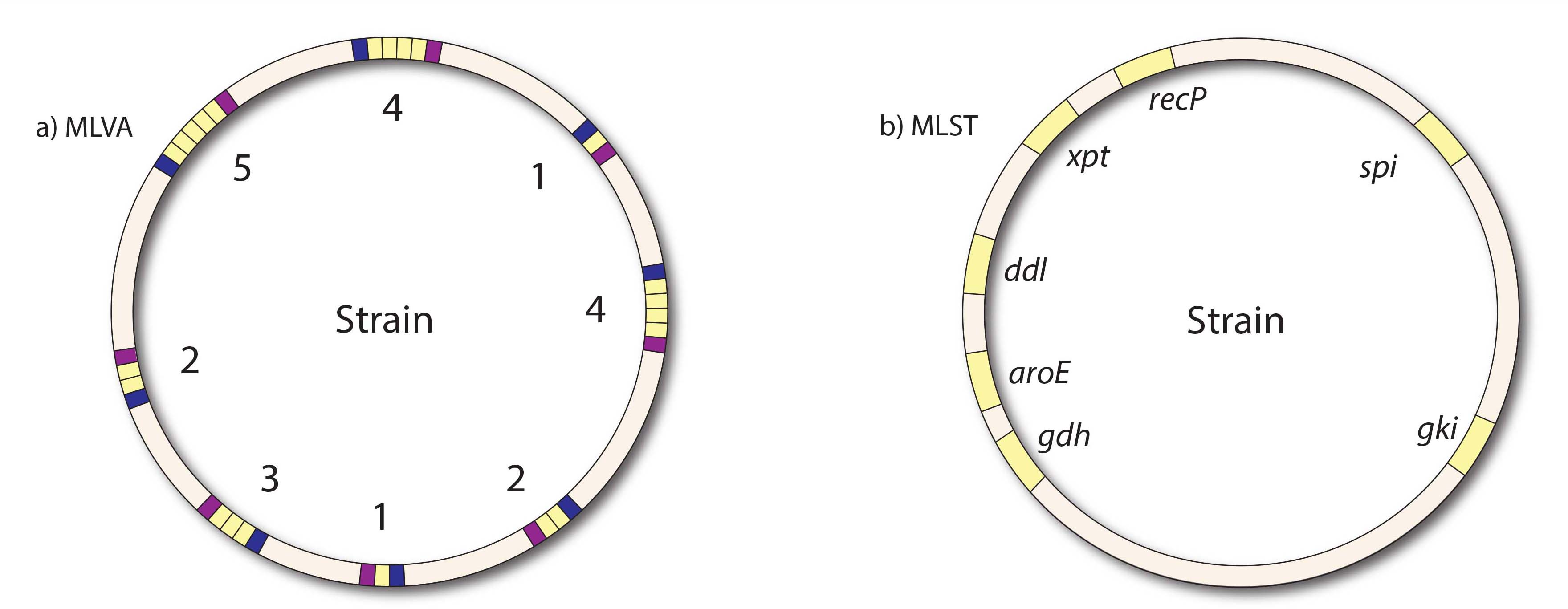
Figure 1. Schematic representation of the MLVA and MLST. a) schematic overview of the MLVA, of 8 loci the number of tandemrepeats are determined. b) Schematic overview of the MLST, partial sequences of 7 housekeeping genes are determined.
Results
In total, the MLST and MLVA yielded 126 and 176 types,
respectively. The MLST and MLVA performed on the Catalan strains
yielded 73 and 92 types, respectively, and on the strains from
the Netherland, 70 and 99 types, respectively. The discriminatory
power of both methods, calculated using the Simpson’s diversity
index (SID), was high (Table 1). Congruence, calculated
using Wallace coefficient, differed per region (Table 2).
|
|
SID |
[95% Cl] |
Number of types |
|---|---|---|---|
|
Serotype |
0.934 |
[0.923 - 0.946] |
39 |
|
MLST |
0.969 |
[0.960 - 0.978] |
126 |
|
MLVA |
0.978 |
[0.970 - 0.986] |
176 |
The Simpsons indices of diversity were high for both methods and comparable between the countries.
|
Spain: |
Serotype |
MLST_ST |
MLVA_MT |
|---|---|---|---|
|
Serotype |
- |
0.458 |
0.314 |
|
MLST |
0.960 |
- |
0.623 |
|
MLVA |
0.961 |
0.935 |
- |
|
|
|
|
|
|
The Netherlands: |
|
|
|
|
Serotype |
- |
0.512 |
0.415 |
|
MLST |
0.985 |
- |
0.482 |
|
MLVA |
0.997 |
0.597 |
- |
The probability of 2 Catalan strains having the same MLVA type
also sharing the same sequence type was 93.5%. However, in the
Dutch strains this probability was only
59.7%.
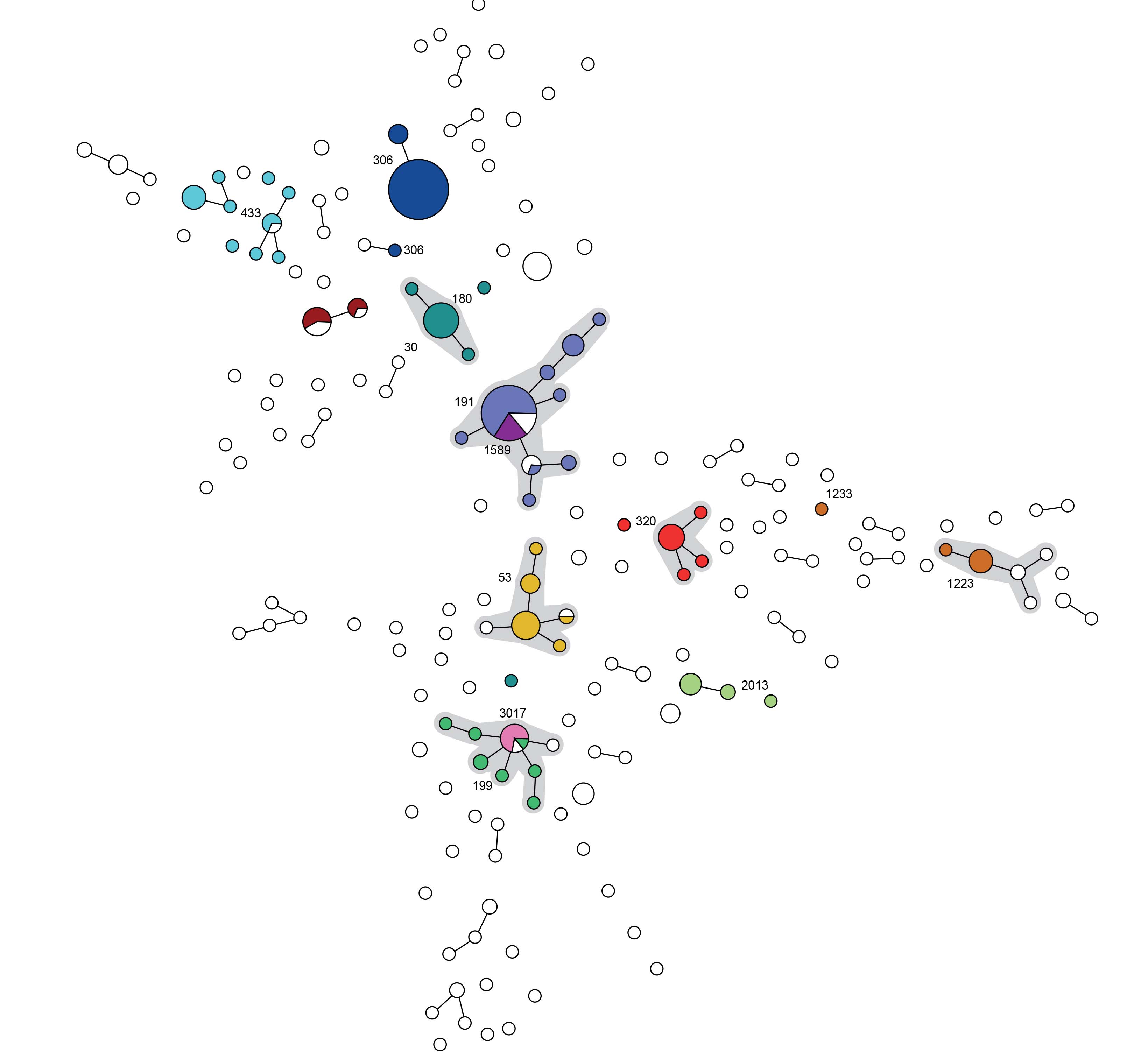
Major clones detected with MLST were ST306 (n=22; 13.5%), ST191 (n=15; 9.2%) and the mul- MLVA 1.000 0.500 - tiresistant clone ST320 (n=10; 6.1%) in Catalonia and ST191 (n=17;10.2%, ST306 (n=15; 9.0%) and ST53 (n=11; 6.6%) in the Netherlands (Figure 2). Difference in clonality of the pneumococcal populations in both regions may provide an explanation for the difference in Wallace coefficient (Table 3).
Figure 2. High congruence between MLVA and MLST. Minimum spanning tree of the results of MLVA (n=329). The colors and text indicate the prevalent Sequence Types within this study.
|
Serotype 19A, Penicillin non-susceptible MIC>0.06 (n=19) |
SID |
[95% Cl] |
Number of types |
|---|---|---|---|
|
MLST |
0.684 |
[0.561 - 0.808] |
4 |
|
MLVA |
0.869 |
[0.790 - 0.948] |
8 |
|
|
Serotype |
MLST |
MLVA |
|
Serotype |
- |
0.316 |
0.131 |
|
MLST |
1.000 |
- |
0.435 |
|
MLVA |
1.000 |
1.000 |
- |
|
|
|
|
|
|
Serotype 19A, Penicillin susceptible (MIC ≤ 0.06) (n=44) |
SID |
[95% Cl] |
Number of types |
|
MLST |
0.890 |
[0.809 - 0.971] |
13 |
|
MLVA |
0.920 |
[0.829 - 1.000] |
17 |
|
|
Serotype |
MLST |
MLVA |
|
Serotype |
- |
0.110 |
0.080 |
|
MLST |
1.000 |
- |
0.344 |
|
MLVA |
1.000 |
0.500 |
- |
The probability of 2
non-susceptible serotype 19A strains having the same MLVA type also
sharing the same sequence type was 100%. However, for susceptible
serotype 19A strains this probability was only 50%. The same holds
true for all susceptible and non-susceptible serotypes analysed
together, but less pronounced. An explanation could be that the
housekeeping genes in non-sceptible strains are more conserved
compared to BOX loci in those strains.
Conclusions
Both methods yield a high diversity index and congruence between the methods was high, but differed between. Using MLVA, we could further distinguish highly clonal isolates, such as the penicillin non-susceptible clones, when grouped by MLST. Although MLST is globally used as gold standard in genotyping of the pneumococcus, the MLVA yield comparable results, may be preferable for typing highly clonal populations and is the cheaper alternative.